Time series processing in R: from raw measurements to their analysis and interpretation
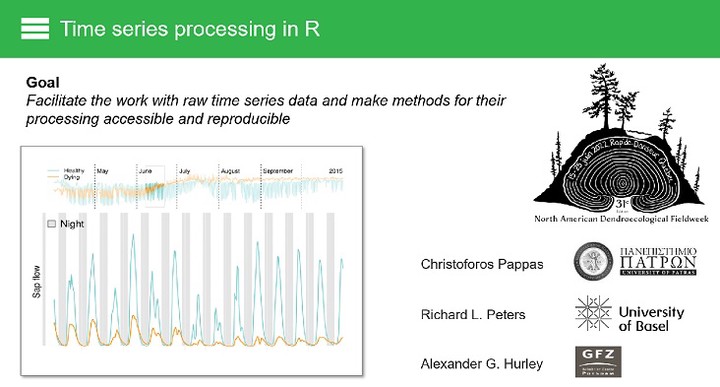 Image credit: C. Pappas
Image credit: C. Pappas
Time series observations provide systematic monitoring of different phenomena, including biogeophysical and environmental processes. However, continuous monitoring can result in rapidly expanding volumes of data, making their handling, exploration, and processing challenging. In this workshop we present workflows for handling time series measurements in the R computational environment. Four key topics are covered: (1) Time series handling, where we present how to import and work with time series data in R, including handling and homogenization of time stamps (e.g., time zones, daylight savings, regular and irregular time steps, etc.) and temporal aggregation (e.g., from minutes to hours, days, etc.). (2) Interactive exploration and reproducible cleaning of time series, where the capabilities of the recently developed ‘datacleanr’ package (https://the-hull.github.io/datacleanr) are used to pinpoint common errors/outliers in various data types. (3) Standardized time series processing workflows for widely used monitoring techniques, such as measurements from automatic stem dendrometers (‘treenetproc’; https://github.com/treenet/treenetproc), sap flow sensors (‘trex’; https://the-hull.github.io/TREX), and quantitative wood anatomy (‘raptor’; https://the-hull.github.io/raptor) are explored. Moreover, additional well-established R packages for processing ecophysiological and biogeophysical time series that are openly available to the community are highlighted. (4) Basics of time series analysis, that includes calculations of descriptive statistics and auto- and cross-correlations, diurnal cycles, hysteresis, and temporal trends, are presented, as well as data interpretations using examples from recent literature to illustrate how the analysis of such time series can reveal new insights and advance our understanding of the underlying biological processes.
The presentation for the course can be found here.
